| Goals: | Illustrate building protactinyl pentahydrate with D5 symmetry. | |
| Demonstrate Symmetry Toolkit operations: Assign Symmetry - Align to Axis - Generate Ghosts - Translate - Generate Molecule - Find Irreducible Fragment. |
|
Prerequisites: You have completed previous examples or already know how to
Setup for Display: To approximate the screen displays in this example, set the following options in the Builder:
| In the Builder | ||
| a. | ||
| b. | ||
| c. | Select View menu, Axes to display the Cartesian axes. | |
| d. | Select Options menu, Auto Normalize Display. | |
| e. | Add a divalent Pa atom (linear sp valence) to the window. | |
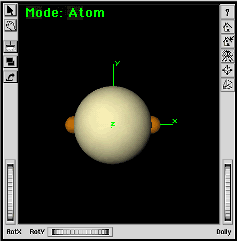 |
The atom will be located at the origin. With the axes displayed, you must click the mouse away from the origin. Otherwise the atom will not be added. | |
| f. | ||
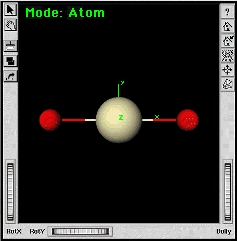 |
You should now have protactinyl (PaO2) in the Builder window. | |
| a. | From the Toolkits menu, choose Symmetry Toolkit to open the Symmetry Toolkit. | |
| b. | Assign D5 symmetry by changing the specification
in the Symmetry Toolkit (not the Builder window). |
|
| c. | |
|
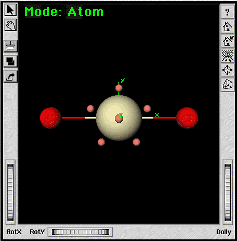 |
The 5-fold symmetry axis is the Z axis by definition. The next step is to line up the O-Pa-O bonds along this axis. | |
| d. | Select the Align to Axis
option in the Symmetry Toolkit. Select the Z button to automatically fill in the parallel axis vector (0,0,1). |
|
| e. | ||
| f. | Turn ON the All option
near the bottom of the Symmetry Toolkit. Select the Align button. |
|
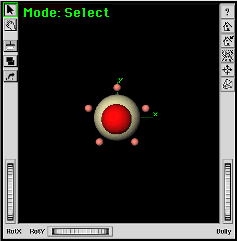 |
The chemical system should realign itself to the Z axis. |
|
| a. | ||
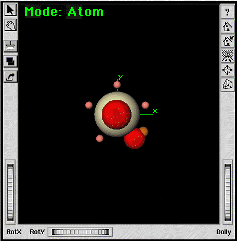 |
The new oxygen atom should be moved so it is 2.6 angstroms from the protactinium atom. We want to put the atom on the positive Y axis so that the bonding nubs are facing away from the origin. To do this, you will use the Translate capability of the Symmetry Toolkit to move the new oxygen to (0, 2.6, 0). | |
| b. | Select the Translate option (radio button) in the Symmetry Toolkit. | |
| c. | ||
| d. | Edit the "n-y:" input box to be 2.6. | |
| e. | Turn OFF the All option in the Symmetry Toolkit. |
|
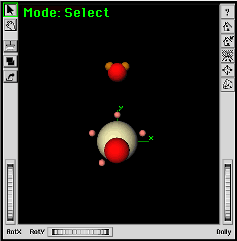 |
The divalent oxygen atom is moved into position on the Y axis. | |
| f. | ||
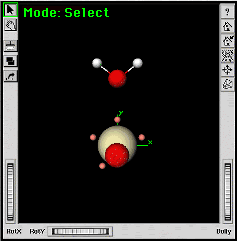 |
||
| g. | ||
| h. | ||
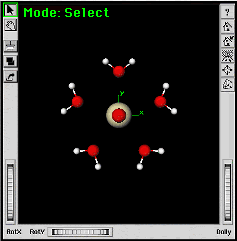 |
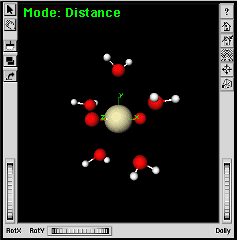
You should have five water molecules around the protactinyl group. | |
| Adjusting the orientation of the water molecules can be done by a series of steps using the Symmetry Toolkit. The initial conformation in this procedure has the H-O-H bond in the XY-plane. Let’s rotate all five water molecules by 90 degrees. | ||
| a. | |
|
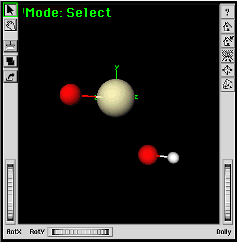 |
You can then use the Torsion Angle Measure/Adjust Tool to adjust the torsion angle between the O-Pa-O-H atoms. The fragment shown here was rotated around the Y axis to show all atoms clearly. |
|
| b. | ||
| c. | While holding the Shift key down on your keyboard, select the four atoms in order, O-Pa-O-H, being sure to select the hydrogen atom last. | |
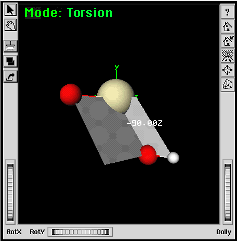 |
Gray planes display to indicate the torsion angle. | |
| d. | Click on the plane displayed in the Builder window to pop open the Torsion window. | |
| e. | 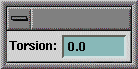 Change
the torsion angle to 0.0 degrees [[and press the Enter key on the keyboard.]] Change
the torsion angle to 0.0 degrees [[and press the Enter key on the keyboard.]] |
|
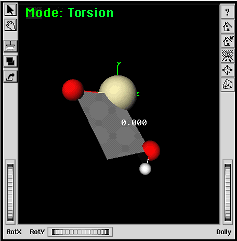 |
The torsion angle changes to 0.0. | |
| f. | ||
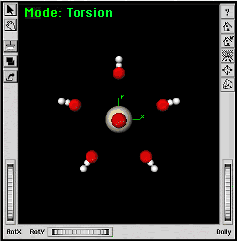 |
The molecular system is reoriented here to show its symmetry. | |
| The default bond lengths for the protactinyl group are too long. These could be adjusted for each bond by using the Geometry Text Table, the Distance Measure/Adjust tool, or the "Translate" option in the Symmetry Toolkit. It is quickest to use the Symmetry Toolkit to reduce the system to unique atoms before making the adjustment. Then only one bond length needs to be modified. | ||
| a. | |
|
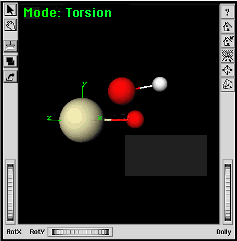 |
You may need to adjust the view so that you can see the Pa-O bond clearly. | |
| b. | ||
| c. | While holding the Shift key down, select the protactinium atom followed by the bonded oxygen atom. | |
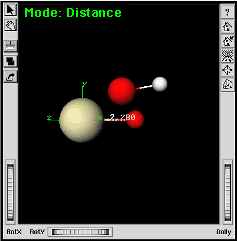 |
The bond length will be displayed. Adjusting this length will now move the oxygen atom. | |
| d. | Click on the Pa-O bond to pop open the Length entry window. | |
| e. | Change the distance to 1.78 angstroms [[and
press the "Enter" key on the keyboard.]] |
|
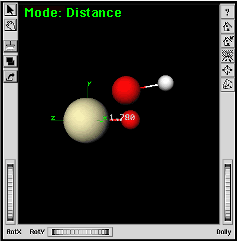 |
||
| f. | ||
 |
||
| Ecce Online Help Revised: August 10, 2003 |
Disclaimer |